ANION EXCHANGE BUFFERS
ANION EXCHANGE BUFFERS
Recommended Buffers for Anion Exchange Chromatography*
To use the calculator, enter the buffer's concentration and temperature, then click on the corresponding button.
The following are the implicit rules of the calculator:
| The best buffers (white background)** | Less suitable buffers (light brown background)** | |
| Ionic strength at at pH=pKa | I ≈ 0.5 C | I ≈ 2 C |
| Concentration range | 1 - 500 mM | 1 - 130 mM |
| Temperature range | 3 - 37oC | 3 - 37oC |
| pKa0 (25 oC) | d(pKa)/dt ΔH0, kJ/mol |
Calculate pKa at given concentration and temperature |
Buffer name/dissociation type | Dissociation step | UV limit |
Notes |
| 5.23 | -0.014 | mM oC pKa |
Pyridine HL+ = H+ + L |
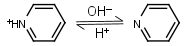 |
275nm | Volatile and toxic. |
| 5.333 | -0.018 31.11 |
mM oC pKa |
Piperazine H2L+2 = H+ + HL+ |
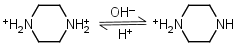 |
Clear | Other pKa: 9.731. |
| 6.05 | -0.017 29.5 |
mM oC pKa |
L-Histidine H2L+ = H+ + HL± |
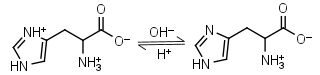 |
235nm | Other pKa: 1.80, 9.34. Chiral. Forms complexes with Me2+, forms complexes with itself. It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 6.484 | -0.017 28.4 |
mM oC pKa |
Bis-tris HL+ = H+ + L |
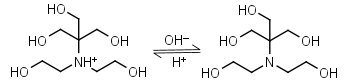 |
Clear | Binds Ca(II), Sr(II), Co(II), Ni(II), Cu(II), Zn(II), Cd(II), Pb(II); weakly binds Mg(II), Ba(II), Mn(II).1 |
| 6.65 | -0.03 |
mM oC pKa |
Bis-tris propane H2L+2 = H+ + HL+ |
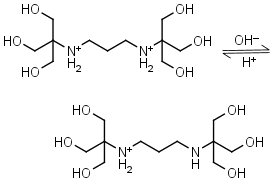 |
Clear | Other pKa: 9.11. |
| 6.993 | -0.021 36.64 |
mM oC pKa |
Imidazole HL+ = H+ + L |
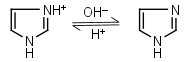 |
235nm | Forms complexes with Me2+, and forms complexes with histidine. Strongly nucleophilic, catalyzes wide range of chemical transformations. |
| 7.77 | -0.016 27.4 |
mM oC pKa |
N-Ethylmorpholine HL+ = H+ + L |
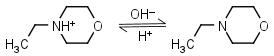 |
Clear | |
| 7.762 | -0.020 33.6 |
mM oC pKa |
TEA (Triethanol- amine) HL+ = H+ + L |
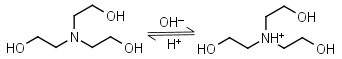 |
Clear | Binds Co(II), Ni(II), Cu(II), Zn(II), Cd(II).1 Can form radicals in the presence of strong oxidants, exercise caution during studies of redox processes. |
| 8.072 | -0.028 47.45 |
mM oC pKa |
Tris HL+ = H+ + L |
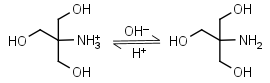 |
Clear | Binds Cu(II), Ni(II).1,2 Binds Co(II), Zn(II), Cd(II), Pb(II); weakly binds Ca(II), Mg(II), Ba(II), Mn(II).1 It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. Inactivates DEPC. Is involved in some enzymatic reactions (e.g. alkaline phosphatase). |
| 8.50 | -0.022 |
mM oC pKa |
Morpholine HL+ = H+ + L |
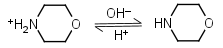 |
Clear | |
| 8.54 | -0.028 |
mM oC pKa |
N-Methyl- diethanolamine HL+ = H+ + L |
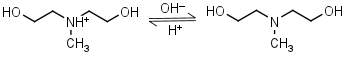 |
Clear | |
| 8.801 | -0.029 49.85 |
mM oC pKa |
AMPD (2-amino-2-methyl-1,3-propanediol) HL+ = H+ + L |
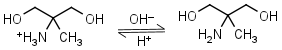 |
Clear | It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 8.883 | -0.026 42.08 |
mM oC pKa |
Diethanolamine HL+ = H+ + L |
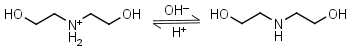 |
Clear | Binds Ni(II), Cu(II), Zn(II), Cd(II).1 |
| 9.498 | -0.030 50.52 |
mM oC pKa |
Ethanolamine HL+ = H+ + L |
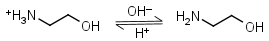 |
Clear | Binds Co(II), Ni(II), Cu(II), Zn(II), Cd(II); weakly binds Mn(II).1 It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 9.694 | -0.032 54.05 |
mM oC pKa |
AMP (2-amino-2-methyl-1-propanol) HL+ = H+ + L |
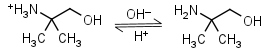 |
Clear | It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 9.731 | -0.025 42.89 |
mM oC pKa |
Piperazine HL+ = H+ + L |
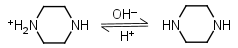 |
Clear | Other pKa: 5.333. |
| 10.55 | -0.026 |
mM oC pKa |
1,3-Diaminopropane HL+ = H+ + L |
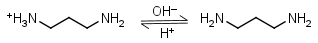 |
Clear | Other pKa: 8.88. Forms strong complexes with many metals. It is a primary amine, and therefore can form Schiff’s bases with aldehydes/ ketones. |
| 11.123 | -0.031 |
mM oC pKa |
Piperidine HL+ = H+ + L |
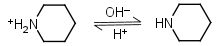 |
Clear |
*
Significant deviations exist in the reported values of pKa and other thermodynamic constants of most common buffers due to them being determined by methods of different accuracy. Additionally, many online resources provide pKa values of biological buffers at unspecified or wrongly specified ionic strengths. We attempted to provide the most consistent data available. pKa0, d(pKa)/dt and ΔH0 are compiled mostly from
- CRC Handbook of Chemistry & Physics, 93th edition: Dissociation Constants of Organic Acids and Bases.
- Goldberg, R. N., Kishore, N., Lennen, R. M. J. Phys. Chem. Ref. Data, 31, 2002, 231-370, as well as some other original publications.
If you have have experimentally observed significantly different values, please report them to support@reachdevices.com.
**Temperature dependence of pKa is presumed to be linear.
The best buffers for cation-exchange (white background):
- for concentrations 1 to 200mM, the Debye-Hückel model is used, and the resulting pKa is presented in brown font.
- for concentrations 201 to 500mM, the Davies model is used, and the resulting pKa is presented in blue font.
Less suitable buffers for cation-exchange (light brown background):
- for concentrations 1 to 50mM, the Debye-Hückel model is used, and the resulting pKa is presented in brown font.
- for concentrations 51 to 130mM, the Davies model is used, and the resulting pKa is presented in blue font.
For a detailed explanation of pKa vs pKa0, and formulas used in the calculator, click on this link.
The buffer is transparent at 200-700 nm range.
1 - Scheller, K. H., Abel, T. H., Polanyi, P. E., Wenk, P. K., Fischer, B. E., Sigel. H. Eur. J. Biochem., 1980, 107, 455-466.
2 - Bai, K. -S., Martell. A. E., J. Inorg. Nucl. Chem., 1969, 31, 1697–1707.
REACH Devices, LLC.
6525 Gunpark Drive, Suite 370-179, Boulder, CO 80301
Call: (720) 288-5722
Inquiries: support@reachdevices.com
Ⓒ 2010-2025 REACH Devices, LLC. All rights reserved.